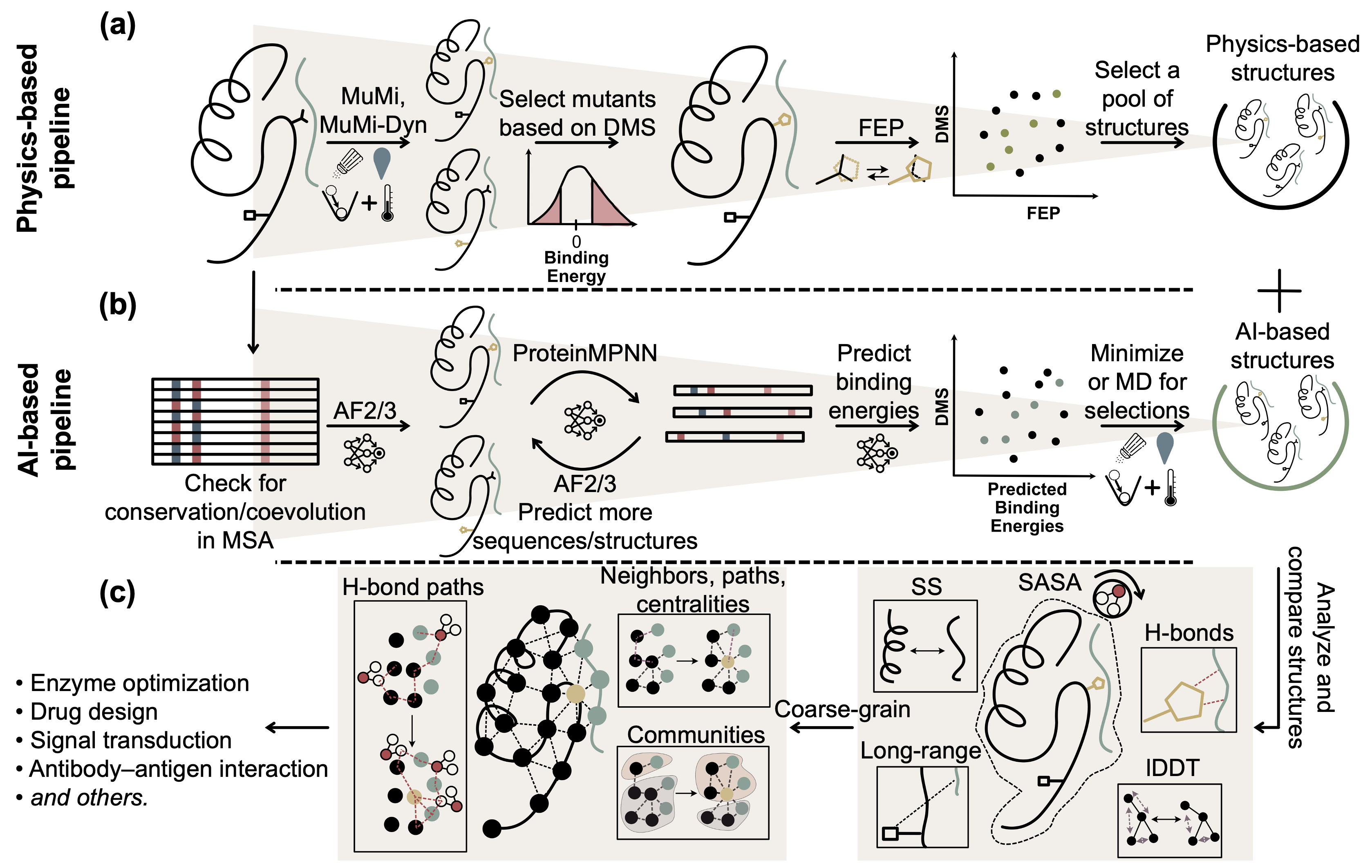
I am an assistant professor at İstinye University. My current focus is on understanding mutational effects in proteins and explaining the molecular basis of deep mutational scanning (DMS) experiments using structural biology. To achieve this, I employ various computational tools including molecular dynamics (MD) simulations, free energy perturbation (FEP) simulations and protein structure prediction tools based on deep learning.
Through MD simulations I investigate the general dynamical features of proteins while FEP simulations allow me to calculate relative binding energies, providing a basis for correlating computational findings with experimental results. I also utilize AlphaFold2 (AF2) for structure prediction; by leveraging multiple sequence alignments (MSAs), AF2 enables prediction of varying conformations through the use of different MSAs.
In analyzing the predicted conformations, I rely on parameters such as hydrogen bonds, solvent-accessible surface area (SASA), relative solvent accessibility (RSA) and distances. By examining probability density functions and variances I aim to detect subtle conformational changes.
Additionally, I use residue networks to scrutinize protein structures. Applying graph theory with metrics like betweenness centrality (BC) and community co-inhabitance (via a BC-based Girvan-Newman algorithm) I aim to explain conformational differences by identifying residues within the same community.
If you are interested in learning more, you can explore my online open lectures:
- Molecular Dynamics Simulations of Small Molecules
- Prediction of Protein Structures Using Deep Learning Tools
- Generating Mutant Protein Structures Using Advanced Techniques
- Residue Networks: Understanding the Time Evolution and Mutations of Proteins from a Graph Theoretical Perspective
Selected Publications
- Guclu T.F., Tayhan B., Cetin E., Atilgan A.R., Atilgan C., High Throughput Mutational Scanning of a Protein via Alchemistry on a High-Performance Computing Resource. Concurrency and Computation: Practice and Experience, 2025, 37: e8371. https://doi.org/10.1002/cpe.8371 – Research Article (Published). (Corresponding Author)
- Guclu T.F., Atilgan A.R., Atilgan C., Deciphering GB1’s Single Mutational Landscape: Insights from MuMi Analysis. The Journal of Physical Chemistry B, 2024, 128 (33), 7987-7996. https://doi.org/10.1021/acs.jpcb.4c04916 – Research Article (Published). (Corresponding Author)
- Guclu T.F., Atilgan A.R., Atilgan C., Dynamic Community Composition Unravels Allosteric Communication in PDZ3. The Journal of Physical Chemistry B, 2021, 125 (9), 2266-2276. https://doi.org/10.1021/acs.jpcb.0c11604 – Research Article (Published).
- Kantarcioglu I., Gaszek I.K., Guclu T.F., Yildiz M.S., Atilgan A.R., Toprak E., Atilgan C., Structural shifts in TolC facilitate Efflux-Mediated β-lactam resistance. Communications Biology, 2024, 7, 1051. https://doi.org/10.1038/s42003-024-06750-0 – Research Article (Published).
- Cetin E., Guclu T.F., Kantarcioglu I., Gaszek I.K., Toprak E., Atilgan A.R., Dedeoglu B., Atilgan C., Kinetic Barrier to Enzyme Inhibition Is Manipulated by Dynamical Local Interactions in E. coli DHFR. Journal of Chemical Information and Modeling, 2023, 63 (15), 4839-4849. https://doi.org/10.1021/acs.jcim.3c00818 – Research Article (Published).
- Guclu T.F., Kocatug N., Atilgan A.R., Atilgan C., N-Terminus of the Third PDZ Domain of PSD-95 Orchestrates Allosteric Communication for Selective Ligand Binding. Journal of Chemical Information and Modeling, 2021, 61 (1), 347-357. https://doi.org/10.1021/acs.jcim.0c01079 – Research Article (Published).
